
Fouling/fish disease link explored
The exact nature of possible links between fouling on netpens, the cleaning of nets and the infection of farmed fish has been the subject of a recent investigation.
The project partners in the pilot study are Steen-Hansen, Uni Research Environment, The Industrial and Aquatic Laboratory (ILAB), Cermaq and the University of Bergen. Financed by the regional research fund for West Norway, the project is investigating which fouling organisms can be important reservoirs for different types of infection. Furthermore, various methods were tried in order to effectively identify different infections in fouling samples. Important questions in this trial were: Which types of fouling organisms can function as infection reservoirs, and which infections (fish pathogens) can survive in the various fouling types?
Methodology
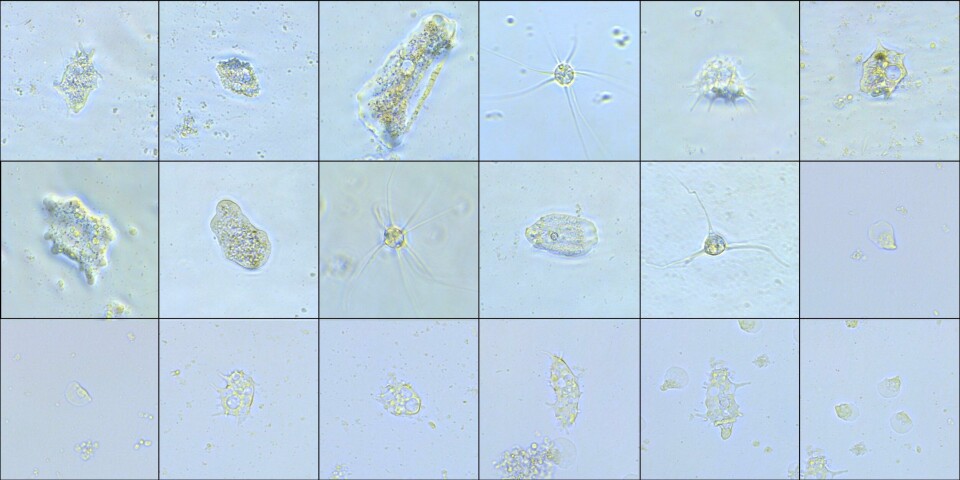
Net frames with fouling from a salmon farm facility were used to carry out an experimental infection trial. The fouling community on the net frames was dominated by hydroids, seaweed, skeleton shrimps and different types of tunicates (sea squirts). There were also settlements of mussels and bush-formed algae. The frames were placed in tanks of seawater (15°C) and a small selection of known fish pathogens that can cause parasite-induced (amoebas) and bacterial diseases were added. Samples of different fouling organisms were taken over a period of 30 days. The trials were used to try out the different molecular test methods (DNA, PCR) for the identification of amoebas and bacteria. In addition, cultivation tests were carried out 10 and 30 days after infection, in order to investigate whether the fouling community still contained living amoebas.
Results
Results from the pilot project show that normal molecular methods adapted to the fish tissue samples and fish pathogens can be insensitive and therefore result in false negatives when analysing fouling organisms to test for the same fish pathogens. Trying out different molecular methods also showed that the challenge of low sensitivity in such analyses can be solved by using methods adapted to the various fouling organisms. These adapted methods increase the sensitivity of the analyses and make it possible to identify just as low levels of infection in fouling organisms as in fish tissue.
The pilot study showed both amoebas and bacteria in all types of fouling, but with a somewhat higher density in seaweed and hydroids. In the cultivation trial a number of different amoeba species were observed 10 and 30 days after the start of the trial. Observed and identified amoebas included the Paramoeba species (the same amoebas that were added in the infection trial), but there were also other unidentified types of amoeba present in the samples. Similar amoebas were also isolated from fish gills (salmon and wrasse) originating from the same facility as the net frames. These unidentified amoebas can therefore represent new species or fish pathogens from salmon and wrasse that have not previously been described.
Further work
Results and experiences from this pilot study will be continued in an extended investigation to develop and optimise methods of analysis for identifying fish pathogens in fouling organisms. Furthermore, different types of amoeba must be isolated from the fouling organisms, identified and tested in order to find out whether they can cause fish disease. The extended study will also investigate the geographic prevalence of fouling organisms that can be important reservoirs for a variety of infections.
The aim of these investigations is to increase knowledge and competence that will contribute to the reduction of fouling, risk of infection and outbreaks of disease on marine fish farming facilities by developing and improving anti-fouling agents in net impregnation, optimising operational routines on aquaculture facilities (handling of nets if infection is identified in the fouling) and ensuring more efficient infection monitoring.























































