Genomic selection doubles trout disease survival
Joint research by the United States Department of Agriculture (USDA) and rainbow trout egg producer Troutlodge has shown that genomic selection can double the prediction accuracy for bacterial cold water disease (BCWD), caused by the bacteria Flavobacterium psychrophilum.
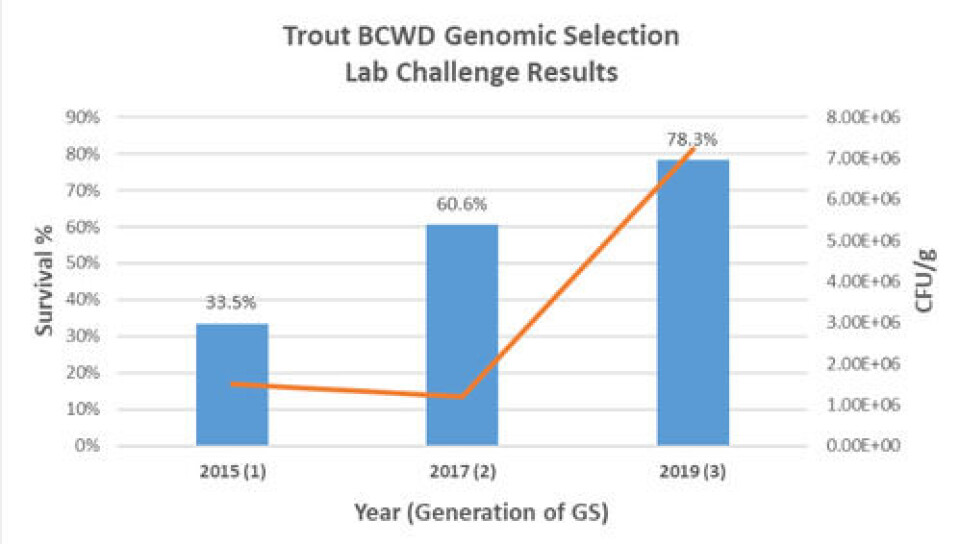
Application of genomic selection for BCWD resistance to three generations of Troutlodge broodstock resulted in a challenge survival increasing from 33.5% in 2015 to 78.3% in 2019 - an improvement of 134%.
Troutlodge, owned by Netherlands company Hendrix Genetics, and the USDA formed their long-term collaboration seven years ago.
SNP chip
The approach included use of genomic selection, made possible when the USDA completed development of a 57K SNP chip in 2014 using Troutlodge genetic stocks for SNP selection and validation.
A single nucleotide polymorphism (SNP) is a variation at a single site in DNA, and the USDA-developed SNP chip, or array, contains 57,000 variations. SNP chips allow rapid testing for specific traits breeders want to include or eliminate in future generations.
The first application of genomic selection to commercial rainbow trout populations took place in 2015, and by 2017 Troutlodge saw an improvement of 80% in survival.
Better resistance
“Since that time, Troutlodge eggs with enhanced survival to BCWD have been sold successfully,” writes senior geneticist Kyle Martin in an article on the company’s website.
“The eggs commercially available today with enhanced resistance to BCWD demonstrate even much better resistance.”
Martin points out that F. psychrophilum is endemic worldwide and impacts both traditional and aquaculture related salmonid production. The pathogen causes outbreaks of both BCWD and rainbow trout fry syndrome, which can cause mortality of up to 85%.
Although a significant amount of research has been done in attempts to develop protection from F. psychrophilum, no effective vaccine has yet been produced, and the molecular pathogenesis involved with infections is not well understood.
Read more here.






















































