

European project provides extensive gene data on top six fish species
AQUA-FAANG ‘has opened the door for precision aquaculture breeding to play a significant role’
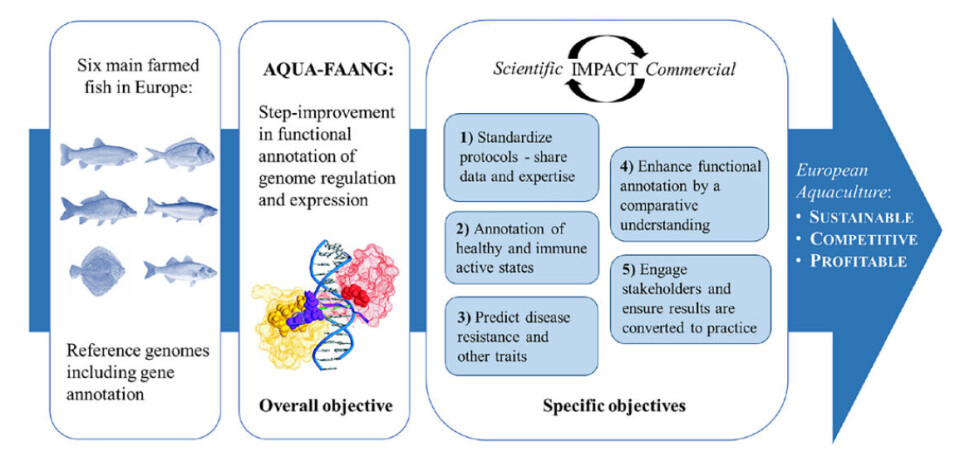
A European project to enhance genomic selection for fish breeding programmes has generated extensive new functional genomics datasets in the six main finfish species farmed in European aquaculture.
The four-year AQUA-FAANG project, supported by the European Union’s Horizon 2020 initiative, involved 25 partners from the UK, Norway, Spain, France, Italy, Greece, the Netherlands, and Poland, and was coordinated by Professor Sigbjørn Lien of the Norwegian University of Life Sciences (NMBU) and Professor Dan Macqueen of the Roslin Institute, Edinburgh University.
Results of the research into the European seabass, gilthead seabream, rainbow trout, Atlantic salmon, common carp, and turbot will provide valuable opportunities to develop precision breeding strategies to increase aquaculture sustainability and profitability.
In an article about the project, Lien and Macqueen write that finfish farming is faces many challenges to sustainable production and expansion, including the constant threat of infectious diseases and climate change, which is enabling new pathogens to enter aquaculture systems while posing further issues to fish health and welfare.
A multi-pronged approach
Addressing such challenges requires a multi-pronged approach, with selective breeding providing a keystone strategy to ensure fish with the most desirable characteristics are farmed now and in the future.
Genomics provides a vital toolbox for commercial breeding programmes in farmed animals. Selective breeding using genetic markers linked to traits (genomic selection - GS) is currently widely used to farm fish with favourable characteristics, e.g., disease resistance.
But while the current generation of selective breeding is effective when the animals are close relatives, it becomes less effective when animals are distantly related from different populations, or across generations, the scientists explain.
This is because the selection performed is not directly based on the underlying genetic variants causing the desired traits.
Precision breeding
“There are millions of genetic variants within any species, but most have zero effect on traits,” Lien and Macqueen write. “If selective breeding involved targeting the much smaller number of genetic variants that truly influence favourable characteristics (causal variants), it would become highly effective in distantly related animals.
“This type of precision breeding would have a range of positive impacts that drive increased sustainability and profitability of the aquaculture sector. However, identifying causal variants is challenging and requires cutting-edge techniques.”
AQUA-FAANG, which ran from May 2019 to May 2023, has produced extensive new datasets to identify regions of the genetic code (i.e., the genome sequence) that impact biological traits in the main finfish species farmed in European aquaculture.
A huge effort
“We call these regions functional, and they include DNA elements that control how genes are expressed in different conditions, for example during development or following a challenge to the immune system,” explain the authors. “Genetic variants in such regions of the genome are much more likely to affect traits of importance to aquaculture than randomly selected variants.
“The AQUA-FAANG project represents a great advancement because this type of data is extremely limited in farmed animals and its generation was a huge effort spanning several years, requiring collaboration by many partner organisations around Europe.”
The AQUA-FAANG approach involves genome functional annotation, leveraging advanced high-throughput DNA sequencing technologies to reveal and describe different types of functional regions in genomes.
Almost 5,000 new datasets
The project generated genome functional annotation data across diverse sample types in the six species, including:
- Different stages of embryonic development – these early life stages play a crucial role in determining traits of relevance to aquaculture and adult fish health.
- Different tissues of adult fishes of both sexes at two stages of sexual maturation – these tissues were selected for relevance to a range of aquaculture traits.
- After immune stimulation with viral and bacterial molecular signals – sampling the primary immune organ of fish.
This comprehensive set of samples was used to generate almost 5,000 new sequencing datasets across the six target species.
Better accuracy
Lien and Macqueen said the AQUA-FAANG datasets add multiple new layers of information on the genetic architecture of farmed fish characteristics, and allow researchers, for the first time, to prioritise genetic variants with increased probability of impacting traits in the target species.
This information can be used in a range of applications, including building new genetic tools and approaches for breeding that have better accuracy without adding significant financial costs.
It can also help reduce the current reliance of industry on measuring important breeding traits (e.g., disease challenge experiments) across different generations, which is expensive and problematic with regard to animal welfare.
Data shared openly
The AQUA-FAANG project shares all its data openly through the Ensembl Genome Browser, which enables researchers and industry to scan the genome of each target species to identify functional regions of interest.
Furthermore, Ensembl unlocks opportunities for researchers and aquaculture breeding companies to quickly and easily overlap their bespoke datasets with AQUA-FAANG annotations, empowering new opportunities for precision breeding applications in different countries and organisations across the globe.
“AQUA-FAANG has opened the door for precision aquaculture breeding to play a significant role in addressing the challenges and opportunities outlined, contributing to the development of a sustainable, resilient, and competitive EU aquaculture sector,” concluded the authors.























































